Your shopping cart is empty!
Setting Up New Online Account and Contact Guideline
Our online ordering system enables direct order of 1st BASE Services such as Standard Sequencing Services, Sequencing+ PLUS Services, Primer Walking and Avian DNA Testing. The setup of online order for 1st BASE and all other products from Axil Scientific will be available soon.
During the transition period, we will transfer the primary email address of active customers as the new login ID into this new online ordering system. You will receive an email notification and step-by-step guideline to smoothen the account information transfer and confirm the new registration. Alternatively, please contact us using LiveChat or WhatsApp during our business hours for direct assistance.
Reward Points on AxillentHub
For every SGD 1 spent, you’ll earn 1 reward point.
You can view your reward points in either of the following ways:
- Click the login icon and select Reward Points
- Go to your Order History page and click Reward Points in the side navigation bar
Visit our Gift Store and select the rewards you’d like to redeem with your points. After adding your chosen items to the cart:
- Check the box “Use Reward Points (Available XXXX)”
- Enter the number of points you’d like to apply
Note: XXXX refers to the total number of reward points currently available in your account.
We currently do not support partial redemption. Please ensure you have sufficient points to redeem your selected items, as redemptions with insufficient points will not be processed. We seek your kind understanding.
Please select Delivery for your redemption. Deliveries will be arranged within approximately 2–4 weeks. Kindly note that we do not deliver to residential addresses.
You can access your cart in either of the following ways:
- Click the login icon and select View Cart
- Go to your Order History page and click View Cart in the side navigation bar
Transition from 1st BASE Online Ordering (1oo) to AxillentHub
The 1oo portal will remain accessible for ordering until 31st July 2025, but all new orders should now be placed through AxillentHub.
Yes, your existing account has been migrated. Just log in with your email address and reset your password if needed.
Yes, you may register a new account at: https://orders.axilscientific.com/register
Please remember to update your mailing address during your first login to ensure a smooth ordering experience.
No, past results will not be accessible on AxillentHub. However, you can still retrieve your previous sequencing results from 1oo until 31st December 2025. We recommend downloading any important data as soon as possible.
Your existing points will remain in the 1oo portal and can be redeemed until 31 December 2025.
You’ll start collecting new points with your first order on the new platform, AxillentHub.
No, the usual collection points at your location remain the same. The zone/building names you see on the portal are for standardisation purposes only. If your building does not have a designated collection point, please keep the samples in your lab, our courier will reach out as per the usual arrangement.
FAQ for Sanger DNA Sequencing Services
We offer trials for new customers to try out our Sanger DNA sequencing services. Please contact our sales representative to find out more.
You will be able to view the price upon ordering on our Online Ordering system. If you are placing an order with us for the first time, the displayed price online might not be updated. Please do not worry as you can still proceed with the ordering. We will update the pricing accordingly as soon as we confirm your order.
1st BASE is the leading brand of Sanger DNA sequencing services, offering high quality results at fast turnaround time. The competitive edge of 1st BASE DNA Sequencing Service includes:
- High quality and long read lengths (~ 1000 bases) for pure DNA template
- Fast turnaround time
- Wide range of free sequencing primers available
- 1st BASE DNA Sequencing Collection Kit available for ease of organizing DNA templates and sequencing primers before sending your order
- Stringent automated processes to include control reaction in each of our processes. Only QC passed sequencing reaction(s) released to our valued customer
- Online ordering system allows monitoring your order status on real-time
FAQ for Sequencing Order Preparation & Submission
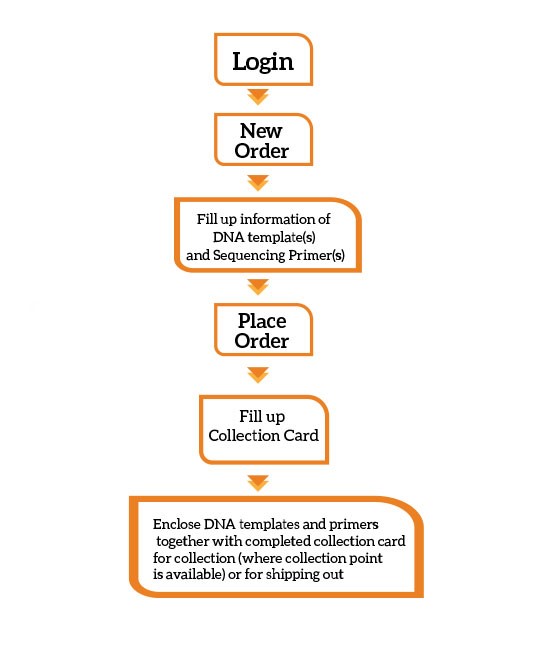
You can place an order with us via online:
Remember to label the Online Order ID on the provided Sample Collection Card or your own packaging. Kindly refer to our quick guide for order registration and submission.
We offer comprehensive service to meet your needs! You can choose the service based on your DNA template type. If you are unsure, kindly contact us.
| Single Pass DNA Sequencing | Standard Sanger DNA sequencing service for purified DNA templates. Guaranteed read length of over 1000 bases for good quality DNA templates. |
| DNA Sequencing+ PLUS | Complete Sanger DNA sequencing solution from unpurified PCR and unpurified plasmid products. Guaranteed read length of over 1000 bases. |
| Ready-to-load Sequencing | Electrophoresis of purified or unpurified cycle sequencing products on Genetic Analyzers. |
| Primer Walking | Includes primer design and synthesis, sequencing with the designed primers, and sequence alignment to generate a consensus sequence. |
1st BASE DNA Sequencing Workflow
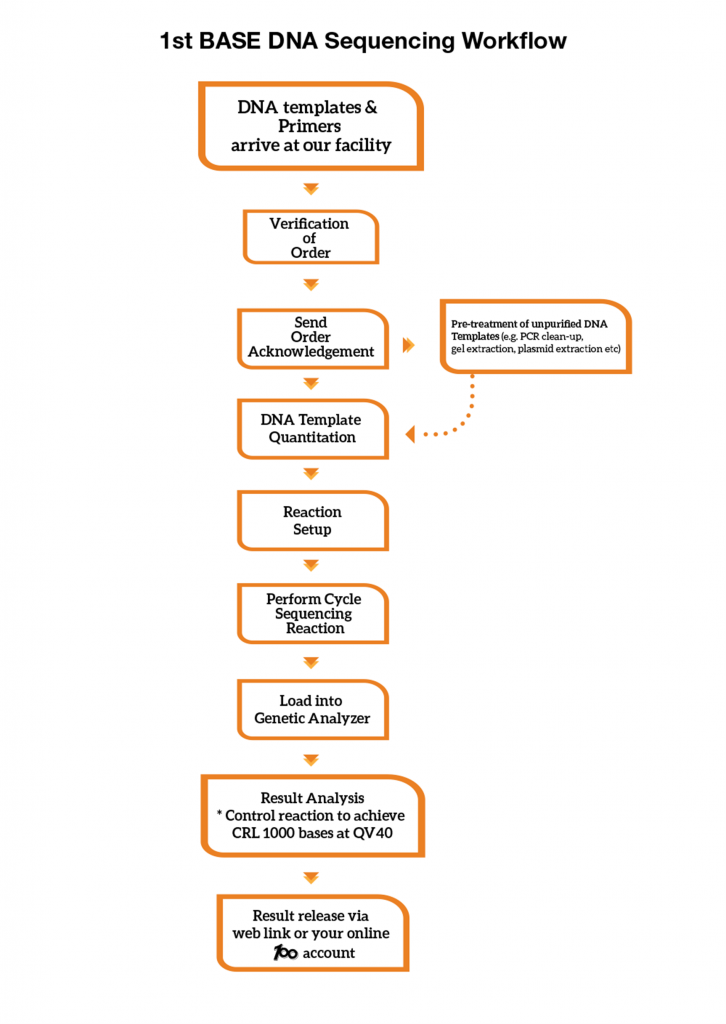
A control sequencing reaction is performed with every run in our Genetic Analyzer. Sequencing reaction controls are used to evaluate the quality of the overall reactions. Our control reaction must achieve Phred40 data and a minimum Contiguous Read Length (CRL) of 1000 bases to meet our Quality Control (QC) benchmark. We provide our control sequencing electropherogram in each of your sequencing order at no charge.
The quality of Sanger sequencing result is closely dependent on the purity and optimum amount of DNA template.
For plasmid DNA:
Most commercial DNA clean-up kits will give DNA of sufficient quality for Sanger sequencing. The most common problem arises from excess salts which usually occur if DNA was eluted in a high salt buffer or diluted/suspended in TE buffers. For Sanger DNA sequencing, we recommend keeping the plasmid DNA in 10mM Tris-HCI pH8.0 buffer or deionized water. TE buffer also can be used to keep the plasmid DNA; but you must pay extra attention for low copy plasmid, where EDTA from the TE buffer may weaken the sequencing signal. Low EDTA TE buffer or TE Buffer with reduced EDTA will be a better option.
Alternatively, we provide plasmid extraction service as an add-on service.
For PCR products:
PCR clean-up is required to ensure that the DNA is free of contaminants, excess PCR primers and dNTPs. Unpurified PCR products are not suitable for direct sequencing as the excess PCR primers will interfere the sequencing signals, which the DNA template supposed to be sequenced by the assigned single sequencing primer for each sequencing reaction. It is also highly recommended that an agarose gel is run to verify that the PCR products is single band with correct target size. Purification can be done using any commercially available kits for gel extraction or PCR clean-up. Alternatively, we provide PCR clean-up and gel extraction service as an add-on service.
Kindly refer our order preparation guideline for details.
If the residues of the PCR primers were depleted after the PCR, the noise background generated during the cycle sequencing may not affect the major base calls. However, the presence of the other contaminants (e.g. dNTPs, salts and etc.) could still compromise the quality of the DNA sequencing result to some extents.
If your unpurified PCR products is single DNA band product on agarose gel, you may order DNA Sequencing + PLUS directly. We will purify the PCR products and enable the remaining DNA to be sequenced immediately in the same sequencing order.
If you wish to verify the unpurified PCR products with only 1 sequencing primer and want the purified PCR products to be sequenced using other sequencing primer in the next new sequencing order, you must order Single Pass Sequencing with add-on PCR clean-up purification service in the first place; so that there will be sufficient purified PCR products to be sequenced in your next new order. Do note that we will only keep the remaining DNA template for 1 week after the results was released. Thus, it will be much straight forward to have all your desired sequencing primers used in the first sequencing order to minimize delays and avoid disappointment.
It is not necessary to lyophilize or dry your DNA templates before Sanger DNA sequencing services. Please submit your DNA templates in deionized water or low EDTA buffer at room temperature. DNA is stable in water at room temperature for a few days.
The quality of DNA template is the most common issue in problematic Sanger DNA sequencing reads. The amount of DNA templates used in sequencing reaction can affect the quality of data. Too little DNA template will generate low signal strength while too much DNA templates will generate a ski-slope effect which causes the signal to weaken gradually.
It is essential to accurately quantitate DNA templates prior to sequencing. However, accurate quantification of dsDNA is not easy.
It’s informative to run your DNA template on an agarose gel, using some standard of known concentration and estimate the relative fluorescent intensity following gel staining. DNA ladders/markers can be used as a reference standard since it comes with a known amount of DNA (ng) for each DNA band.
Kindly refer our guidelines to read the agarose gel electrophoresis for details.
The regular gel photo shall display different DNA intensity for its known DNA Ladder, so that we can use it to estimate the DNA amount of your purified PCR products that loaded next to the DNA Ladder.
By using the example of gel photos below, when we compare the DNA intensity of the purified PCR Products with its DNA Ladder: the known DNA size of 600bp (from DNA Ladder) near to the target PCR products is 12ng and it looks brighter, we can thus confidently say that the purified PCR Products has < 12ng of DNA. If we load only 1µL of purified PCR Products in this agarose gel, we can then estimate the DNA concentration of the purified PCR Products is < 12ng/ µL, which you can take any number between 5ng/µL to 9ng/µL, but not any figure near 12ng/µL.
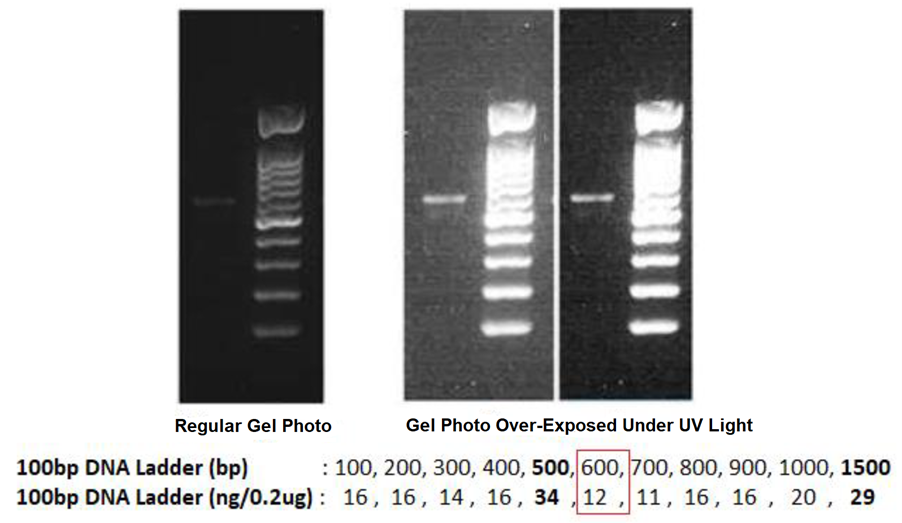
If the agarose gel is over-exposed under the UV light during the gel photo is taken by any gel documentation system, the displayed intensity of the DNA ladder will not be able to guide us to estimate the DNA amount confidently. Although the DNA intensity of the purified PCR products is higher in this gel photo, but the actual DNA concentration is only 5ng/µL to 9ng/µL under regular gel photo.
Generally, there are 2 commonly used methods to quantitate DNA templates. But neither method will show the presence of contaminating salts.
-
- Agarose gel electrophoresis – Purified DNA should run as a single band on an agarose gel (Uncut plasmid DNA can be visualized as 3 bands: supercoiled, nicked, and liner). Contaminating DNA or RNA will show up in the gel.
- UV Spectrophotometry - The A260/A280 ratio should be between 1.7 and 1.9. Smaller ratios usually indicate the presence of contaminating proteins or phenol. The NanoDrop ND-1000 spectrophotometer takes 1 to 2 µL of sample and can detect concentrations from 2 to 3700 ng/µl without dilutions.
Traditional absorbance invariably overestimates DNA. In a finding, Kapp et al (2014) encounter a five-fold difference between absorbance-based method and fluorescent-based method, which could theoretically result in using lower amounts of starting DNA for the downstream sequencing step and cause unsatisfactory results. Discrepancies between the two methods were also reported by Deben et al, O'Neill et al and Simbolo et al (2013). The measurements at 260nm (A260) fluctuate with contaminants and buffer component, mainly because miniprep kits on the market often inevitably introduce some RNA, chromosomal DNA, and other fluorescent cellular material during the purification process that will absorb UV light. These other chemicals won’t necessarily inhibit the sequencing reaction, but they will contribute to the A260 reading. As a result, relying on the spectrophotometer reading alone will cause you to add less DNA template than you think.
Fluorescence assays on the other hand are less prone to interference. Fluorescence-based quantitation is more sensitive and more specific for the nucleic acid of interest. In the technical note from Invitrogen, comparison between the UV absorbance method and the fluorescent method was compared. Nanodrop method was found to shown fluctuation particularly at lower concentration range. Whereas, fluorescence-based quantitation generate accurate and precise result across lower concentration range.
Our laboratories adopt fluorescence-based measurement which might explain the difference in the measured DNA concentration. We take every step to ensure that the quantification process is consistent and accurate for downstream processes. Our laboratory apparatus is calibrated and maintained regularly to ensure they are working within their specified limits.
References:
Kapp JR, Diss T, Spicer J, et al Variation in pre-PCR processing of FFPE samples leads to discrepancies in BRAF and EGFR mutation detection: a diagnostic RING trial Journal of Clinical Pathology Published Online First: 27 November 2014. doi: 10.1136/jclinpath-2014-202644
Simbolo M, Gottardi M, Corbo V, et al. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS ONE 2013;8:e62692
Invitrogen™ Technical Note. Comparison of fluorescence-based quantitation with UV absorbance measurements: Qubit™ Quantitation Platform vs. spectrophotometer
We do not recommend sending DNA template of insufficient amount. If you request us to process such DNA template to sequencing, we will try to deliver our best effort guarantee to maximize the success of your sequencing order. Although a specialized protocol will be deployed to maximize the amplification (and there are just so much we can do), we are still unable to guarantee optimal DNA sequencing result.
Ideally DNA templates should be 100bp and above. With our optimized running protocol and module, we can sequence DNA templates as small as 90-100bp, but this will be subjected to the DNA template quality and the priming efficiency of its sequencing primer.
For DNA templates less than 90bp, we recommend the sequencing to be done using Next-Generation Sequencing.
We will only keep DNA templates and sequencing primers for 1 week upon receiving them. Please indicate in the order form if you would like us to keep your DNA templates or sequencing primers for further sequencing reactions. A nominal fee will apply for storing your DNA templates or sequencing primers in our laboratories.
FAQ for Sequencing Results
Upon the receipt of your sequencing order to our DNA sequencing facility, an order confirmation will be sent to you through email. We typically deliver the sequencing result to you within 48 working hours upon the receipt of your DNA sequencing order with complete ordering information. For ready-to-load reactions, results are typically ready within 24 working hours.
Kindly refer our list of DNA sequencing services for details.
Results will be downloaded through Online Ordering account. The compressed file in .zip format (for Windows), contains the complete sequencing results with (i) electropherogram(s) in .ab1 file and (ii) fasta sequence in .seq file.
We understand that the confidentiality of your result data is critical. To ensure a high level of security towards the result uploaded, for customers who yet to register an online account with us, we would provide result in email with the weblink that is password protected. The password will only reveal to the email recipient.
To view the electropherogram that saved as .ab1 file, we recommend freeware from Applied Biosystems® Sequence Scanner 2.0 for Windows 10 or above, which enable automatic Quality Control Report at a glance. Anyhow, this file can also be viewed by other freeware like Chromas, FinchTV, and etc.
To view the fasta sequence that saved as .seq file, Notepad or Text can be used.
The results stored in the online account has expiry of 12 months. For customers who do not have the online account, the web link provided in the email to download the results has a validity of 3 months.
Please contact us through LiveChat or email by indicating your online sequencing order ID, so that we could assist you directly.
FAQ for DNA Sequencing Primers
Ideally, sequencing primer should be designed 100 bases upstream of your sequence of interest. Below are some other consideration when doing your primer design for sequencing:
- Length of primer to be between 18-25 bases
- GC content to be between 40-60%
- Melting temperature at around 55-60°C
- Check for significant hairpins (>3bp) and homodimerization
- Choose your sequencing primer desalted from Integrated DNA Technologies (IDT) or HPLC grade from other manufacturers
Many commercial oligo providers offer online design tools which you can use. We recommend IDT’s OligoDesigner.
DNA Sequencing Results Interpretation
& Troubleshooting
For .seq file, it is the nucleotide fasta sequences extracted from electropherogram, and can view by Notepad or Text.
For .ab1 file, it is the electropherogram or sequencing trace file that requires specific software to view. We highly recommend using Applied Biosystems® Sequence Scanner 2.0 software to view any Sanger electropherogram or sequencing trace file. The weblink to download this freeware is provided with each release of 1st BASE DNA Sequencing Order. There is a minimum system requirement to install this software completely. The installation window will check your computer system during installation. Alternatively, the electropherogram can also be viewed using Chromas, FinchTV, DNA Baser and etc.
FAQ for Sample Preparation Services
We have in-house expertise in processing samples for downstream molecular biology applications routinely. Researchers may depend on us to process their samples for their processes without fuss.
The process of sample preparation is tedious and the amount of samples are typically limited for research. To maximise success for each sample preparation service, we advise our customers on how to collect and submit the samples within the acceptable terms and conditions. Such information is also available in each of the respective order forms. Minimum setup fee will apply when sample preparation service fails due to bad quality of sample that did not meet the prescribed terms and conditions.
**Note: For raw materials not within our submission guidelines, please download Order Form: Sample Preparation Services for Non-standard Raw Material Submissions.
We have provided a clear step-by-step guide at the ordering page for Sample Preparation submissions. Download and complete the Biosafety Declaration Form and the Order Form. You may upload the completed forms using the upload buttons provided on the portal. Customers are strongly encouraged to include clear images of your sample as well. There is also a link on the ordering page to view our Sample Preparation Guideline for requirements and recommendations.
You may click on this link to view our Sample Preparation Guideline for requirements and recommendations. If your sample type is not listed within our sample preparation guidelines, kindly contact us directly for more information.
FAQ for Avian DNA Testing
Please read our sample preparation guidelines for Avian DNA Testing before place the order online.
There are three different local language available to ease your understanding.
Our Avian DNA Sexing Test is designed to determine the gender from all kinds of non-ratite birds. Parrot is non-ratite birds and thus its gender can be determined through our designed molecular based DNA testing services.
The Avian DNA Test Report will be downloaded through your Online Ordering account. It contains the result interpretation and printable version of Bird DNA Gender Card(s). No hard copy results will be delivered.


